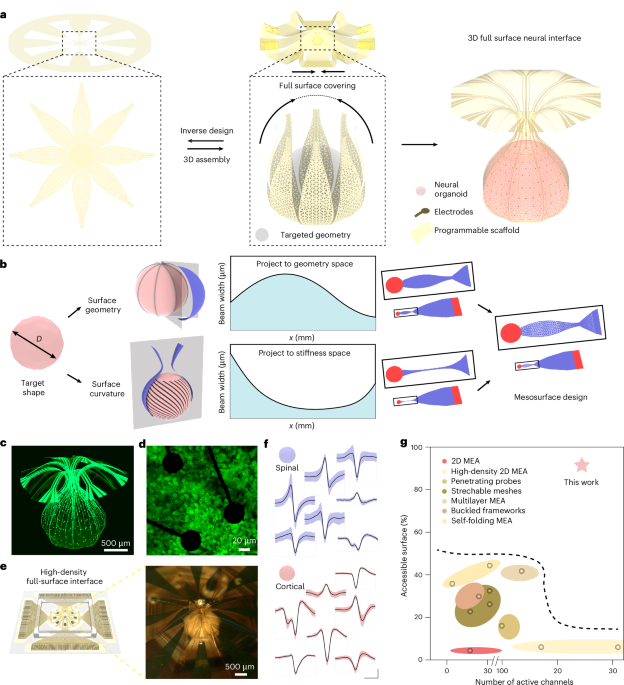
Inverse design strategies
Inverse design used an AGA to determine 2D microlattice patterns, defined by spatially varying triangular voids, for targeted 3D geometries (Extended Data Fig. 1). The optimization process used the Python framework DEAP, a common package for the deployment of AGAs. Discretization of the surface of the target geometry into a finite number of segments formed 2D shapes with width distributions (wg) for continuous surface coverage. These geometries served as the initial population for optimization of the stiffness distribution (ws) through AGA. For each generation, Python codes determined coordinate errors between simulated 3D shapes (ABAQUS) created by the buckling-guided assembly process and the targeted geometry. A set of 50 individuals per generation ensured an accurate determination of ws, defined by an error below a threshold value. The resulting geometries and stiffness distributions defined 2D precursor structures in microlattice layouts of triangular voids, for mechanically guided assembly into the desired 3D configurations.
3D assembly
Fabrication of the 2D precursors involved the following process steps (Supplementary Fig. 1). (1) Spin coating (3,000 rpm for 40 s) and baking (180 °C for 150 s) of poly(methyl methacrylate) on a substrate of glass or a silicon wafer formed a sacrificial release layer. (2) Spin coating (750 rpm for 60 s), baking (120 °C for 600 s, 150 °C for 300 s, 180 °C for 300 s) and vacuum baking (260 °C for 1 h) of a precursor to PI (PI-2545, HD Microsystems) defined the first layer of the polymer scaffold. (3) Spin coating (5,000 rpm for 40 s) and baking (110 °C for 60 s) formed a layer of negative photoresist (AZ nLOF 2035). (4) Photolithography with a maskless aligner (MLA150, Heidelberg Instruments) defined patterns for microelectrodes and interconnects, by 375-nm ultraviolet exposure with a dose of 140 mJ mm−2, followed by post baking (110 °C for 90 s) and developing (immersion in AZ 300MIF for 55 s). (5) Electron beam evaporation yielded a uniform 10-nm Cr adhesion layer and a 200-nm Au film. (6) Lift-off by immersion in a photoresist stripper (AZ 400T) overnight resulted in corresponding patterns of Au/Cr. This step defines the microelectrodes, traces and reference electrodes. (7) Repeating step 2 yielded the second layer of the polymer scaffold. (8) Repeating steps 3 and 4 formed a pattern of photoresist to define the 2D outlines of the polymer scaffold. (9) Thermal evaporation (Denton Vacuum Explorer 14) formed a 50-nm Cu film. (10) The lift-off process resulted in six defined Cu patterns that served as hard masks for plasma etching. (11) Plasma dry etching transferred this Cu pattern into the underlying polymer to form the scaffolds and to expose the microelectrodes. (12) Wet etching removed the remaining Cu, thereby completing the fabrication.
The mechanically guided process for a 3D framework involved the following sequence. (1) Immersion in acetone dissolved the poly(methyl methacrylate) sacrificial layer and released the 2D precursor from the supporting substrate. (2) A piece of polyvinyl alcohol tape allowed retrieval of the precursor and exposure of its backside for sputter deposition of 30-nm SiO2 through a shadow mask, to define the selective bonding sites. (3) Exposure to ultraviolet ozone for 5 min followed by baking at 75 °C for 15 min enabled strong chemical bonds to form between the bonding sites and the surface of a prestretched substrate of PDMS (SYLGARD 184 Silicone Elastomer, with 20:1 mixing ratio) upon physical contact. (4) Release of the prestretch caused the 2D precursor to buckle into a corresponding 3D geometry.
Mechanical analysis
Three-dimensional FEA quantified all aspects of the post-buckling responses and the maximum principal stress/strain distributions in the 3D structures. Four-node 2D shell elements (S4R) modelled the polymer scaffolds, and eight-node 3D solid elements (C3D8R) captured the elastomeric substrates. Refined meshes ensured computational accuracy and efficiency, conducted with commercial software (ABAQUS). Linear elastic constitutive models described the properties of the metal (Au) and polymer (PI), where the elastic modulus and Poisson’s ratio are EAu = 70 GPa and νAu = 0.44; EPI = 2.5 GPa and νPI = 0.35. Modelling of the prestretched silicone substrate relied on an isotropic hyperelastic structure using the Mooney–Rivlin law (EPDMS = 2 MPa and νPDMS = 0.49). The organoids were modelled as an incompressible solid material with an elastic modulus of ~1 kPa. Applying linear buckling analyses for the 2D pattern determined the critical buckling strain and the corresponding lowest buckling mode, which were then considered as initial geometric imperfections in the post-buckling simulations to achieve the final deformed configurations and stress/strain distributions. Notably, the geometric nonlinearity was accounted for in the post-buckling analyses of the 3D configuration.
Electrical characterization
A three-electrode configuration with a Pt wire as the counter electrode and Ag/AgCl as the reference electrode enabled electroplating of Pt black (Autolab PGSTAT128N, Metrohm AG, -0.1 V for 30 s) with a solution of 3 wt% of chloroplatinic acid and 0.1 wt% of lead acetate. A similar three-electrode configuration in phosphate-buffered saline (PBS) enabled electrochemical impedance spectroscopy and charge injection limit measurements on each Pt-coated microelectrode. Measurements of charge injection limits employed biphasic pulses (a cathodic pulse followed by anodic pulse) with a pulse duration of 200 µs. A 2-ms interval between the two pulses defined the access voltage (Va) as the voltage drop at the end of each pulse that originates from ionic conductivity of the electrolyte. The highest (lowest) voltage minus (plus) the access voltage yielded the most positive (negative) voltage, Emax (Emin), as plotted with an example case in Extended Data Fig. 3d. Gradually increasing the pulse current until either Emax = 0.9 V or Emin = −0.6 V defined the maximum charge injection without exceeding the water window. Long-term monitoring of the 3D structures immersed in 37 °C PBS used a commercial system (Intan RHS system, Intan Technologies) to track changes in electrochemical impedance and noise levels for each channel.
Generation of human cortical organoids
This study used human cortical organoids derived from WTC-11 (Coriell, GM25256), BJFF.6 (RRID: CVCL_VU02, Genome Engineering & Stem Cell Center (GESC), Washington University School of Medicine) and AN1.1 (Tech ID: T-019532, GESC, Washington University School of Medicine) hiPS cell lines. The genome integrity of all lines are confirmed by karyotyping (Supplementary Figs. 21, 23 and 24). The process began with culturing cells in mTeSR Plus medium (StemCell Technologies, 08620) plus 1% penicillin and streptomycin (Thermofisher, 15140122) in six-well plates coated with Matrigel (Sigma-Aldrich, CLS354277) for 5 days, followed by dissociation into single-cell suspensions using gentle cell dissociation reagent (StemCell Technologies, 100-0485) and centrifugation at 300g for 5 min. Dorsal Forebrain Organoid Kits (StemCell Technologies, 08620) formed the organoids following a previously published protocol25. In brief, hiPS cells formed uniform aggregates after transfer to Aggrewell 800 microwell culture plates (StemCell Technologies, 34815) at a density of 10,000 cells per microwell. Treatment of the aggregates involved exposure to dual SMAD inhibition for 6 days and with partial daily media changes. On day 7, transfer occurred to ultralow-attachment six-well plates (30–40 organoids per well), supplied with serum-free media containing fibroblast growth factor-2 (FGF-2) and epidermal growth factor (EGF) until day 25. They were then transferred to media containing brain-derived neurotrophic factor and neurotrophic factor 3 until day 43, after which the media was supplied without growth factors. A mixture of four small molecules—GSK2879552 (Selleck Chemicals, S7796), EPZ5676 (Selleck Chemicals, S7062), Bay K 8644 (Selleck Chemicals, S7924) and NMDA (Selleck Chemicals, S7072)—each at a concentration of 1 mM, was added to the media according to a previously published protocol43 to enhance neuron maturation into cortical organoids from day 15 until the day of experiments.
Generation of human spinal organoids
Generation of human spinal organoids (hSCOs) followed an established protocol that was similar to that of the cortical case37, but with differences in the supplements. On day 1, cells (HUES 3 line) (genome integrity confirmed by karyotyping; Supplementary Fig. 22) were cultured with mTeSR Plus Medium (StemCell Technologies). Dissociation of cells using Accutase (Fisher Scientific, 50-112-9055), followed by centrifugation and seeding into the AggreWell 800 (StemCell Technologies, 34815) plates at 10,000 cells per microwell, initiated the aggregation. After a centrifugation at 100g for 3 min, incubation of the plates occurred for 24 h at 37 °C with 5% CO2. From day 0 to day 6, organoid culture was in MTeSR plus medium supplemented with dorsomorphin (2.5 μM, Sigma-Aldrich, P5499) and SB-431542 (10 μM, DNSK, KI-12) with daily media changes, then replaced on day 7 with Neurobasal-A medium containing N-2 (Life Technologies, 17502048), B-27 (50×, minus vitamin A, Gibco, 12-587-001), CHIR (3 μM, MTocris, 4423409), retinoic acid (0.1 μM; Sigma-Aldrich, R2625) and EGF (20 ng ml−1; R&D Systems, 236-EG-200) as growth factors for neural induction. From day 19, the medium was enriched with BDNF (20 ng ml−1, R&D Systems, 248-BDB-250/CF), IGF-1 (10 ng ml−1, Peprotech, 100-11), DAPT (2.5 μM; Tocris, 2634/10), N-2 (Life Technologies, 17502048), L-ascorbic acid (AA, 200 nM; Sigma-Aldrich, A4403) and cAMP (50 nM; Sigma-Aldrich, D0627) to promote further neural maturation. The feeding schedule was every other day until day 41 and subsequently every 4–5 days after day 43 without growth factors.
Immunofluorescence and imaging
Fixation of the organoids occurred through overnight immersion in 4% paraformaldehyde (Thermofisher, J61899.AK) at 4 °C, followed by rinsing three times with Dulbecco’s PBS (DPBS; Thermofisher, 14190136) and storage in PBS (Thermofisher, 10-010-031) at 4 °C. Organoids were transferred to 30% sucrose/PBS solution for 24–48 h before sectioning and placing in cryomolds filled with OCT (Leica Biosystems, 3801480). Cryostat sectioning of samples occurred sequentially, at 14 µm thickness, using a Microm HM525 Cryostat. The sections were then placed on microscope glass slides and stored at −20 °C for subsequent immunostaining.
Immunofluorescence began with DPBS washing of the slides before incubation for 1 h at room temperature in blocking solution (1% BSA (Thermofisher, 15260-037) and 0.1% Triton-X (Sigma-Aldrich, 9036-19-5 diluted in PBS), and then followed by incubation with primary antibodies diluted in the blocking solution overnight at 4 °C. Primary antibodies used in this study included mouse anti-S100B (Sigma-Aldrich, S2532, 1:100), guinea pig anti-MAP2 (SYSY, 188 004, 1:100), rabbit anti-SOX2 (Sigma-Aldrich, AB5603, 1:100), PSD95 (Abcam, ab18258, 5 μg ml−1) and Synaptotagmin-1 (Developmental Studies Hybridoma Bank (DSHB), AB_2199314, 1:100). Subsequently, after washing of the primary antibodies with three 5-min cycles of DPBS, incubation with secondary antibodies occurred at room temperature and protected from light. The secondary antibodies used in this study were Alexa Fluor 488 goat anti-guinea pig (Jackson, 106-545-003; 1:500), Alexa Fluor 647 goat anti-mouse (Jackson, 115-605-003; 1:500), Alexa Fluor 568 goat anti-rabbit (Thermo Fisher Scientific, A-11011), Alexa Fluor 647 goat anti-rabbit (Jackson, 111-605-144; 1:200) and Alexa Fluor 488 goat anti-mouse (Jackson, 115-545-205; 1:200). Adding 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, 10236276001, 1:10,000) for 5 min yielded labelled nuclei. Air drying and placing the slices on a cover slip with mounting medium (Vector Labs, H-1700-10) prepared them for fluorescent imaging with a FV3000 Olympus confocal microscope. Laser settings and exposure times were consistent for each channel throughout all samples.
Integration of organoids with 3D interfaces
Insertion typically happened after day 60 of differentiation for hCOs and day 40 for hSOs. Sterilization of the 3D structure occurred through autoclaving, followed by rinsing with PBS immediately before integration. Placing the interface on a plastic ridge and applying gentle pressure opened the 3D interfaces, allowing insertion of organoids using wide-bore pipettes. Integrated organoids were kept at 37 °C with 5% CO2, with media changes every 2–3 days to support the grow-in-place process before electrophysiological recordings.
Simultaneous electrophysiological and fluorescence recordings and data analysis
Simultaneous recordings of electrophysiology and calcium fluorescence occurred on a confocal microscope. Application of a semi-permanent fluorescent calcium reporter, GCaMP6f (Addgene, 100837-AAV1), induced GFP fluorescence to allow measurements of calcium dynamics by live imaging. Organoids were transduced with adeno-associated virus particles at a 5 × 105 multiplicity of infection and incubated for 72 h. After removing the viral particles, GCaMP6f expressed in 3–5 days. A commercial system (Intan RHD, Intan Technologies) with multiple headstages connected in parallel enabled high-channel number recordings. The Intan RHX software recorded and stored real-time electrophysiological potentials at a sampling rate of 20 kHz, with a bandwidth of 1–9,000 Hz and a notch filter at 60 Hz. Each recording session concluded in less than 5 min to minimize the effects of cooling on organoid activities during the measurements.
The software package ImageJ with a customized Java code served as the basis for evaluating calcium fluorescence images and videos. Each frame was discretized into 100 × 100 units to quantify brightness at each location. Normalizing these brightness data with the average brightness of a baseline image of the same session revealed the spatiotemporal characteristics of organoid activity. Locally weighted scatterplot smoothing with frac = 0.5 in range [0:1] to reduce effects of environmental noise and random fluctuations preceded filtering with a Savitzky–Golay filter with window length = 9, polyorder = 8. Computations of the differences of the normalized brightness between the unit matrix of two imaging frames formed a 2D heatmap map to indicate the 2D distribution of activities.
Data analysis of neural activities used a customized Python code (Extended Data Fig. 5) that followed a multistep procedure. Conversion of the raw data into .npy format preceded processing with a third-order Butterworth bandpass filter between 250 Hz and 3,000 Hz and application of a peak detection algorithm. These latter processes involved calculating the r.m.s. noise of the recording channels and detecting peaks with amplitudes larger than five times of the r.m.s. Defining a 5-ms minimum interval between any two labelled peaks prevented duplication in peak counting. Segmentation of each detected peak by a 3-ms window (60 data points of neural potential for 20 kHz sampling rate) formed a high-dimensional vector. Use of an unsupervised clustering algorithm two-component uniform manifold approximation and projection reduced the dimension of the 60-dimensional vector to 2. Subsequently, density-based spatial clustering of applications with noise (DBSCAN) categorized these 2D vectors into clusters of neural spikes.
Calculations of the average potential and standard deviations (s.d.) of all the neural spikes within each cluster defined the overall characteristic waveforms. Such waveforms collected throughout all recording sessions formed a comprehensive dataset, including both neural activities and artefacts. Systematic recordings with control groups of empty interfaces and interfaces integrated with fixed organoids marked the waveforms associated with mechanical and optical noise in the environment. Excluding spike clusters with these artefact waveforms from the recording data ensured artefact-free raster plots of neural activity.
Additional calculations of the basic characteristics included firing rates and spike amplitudes of each individual channel. Associating the data of each channel with the designed 3D coordinates of each corresponding microelectrode resulted in a 3D spatial cloud data distributed across the surface of a sphere that mimics the geometry of the organoid. A radial basis function (scipy.interpolate.Rbf) then interpolated this cloud data onto a mesh on the spherical surface with points uniformly distributed in spherical coordinates (600 grids in θ and 300 grids in φ). Three-dimensional cross-correlation analysis used pairwise cross-correlations among all recorded neural spike clusters. For the spike times of each pair of recorded clusters, cross-correlograms (CCGs) with a specified range of −200 to 200 ms and a bin width of 0.5 ms followed by boxcar-filtering of 3 ms defined histograms of the amplitudes of delay correlations as a function of time delay. CCG of shuffled spike trains of each cluster in the pair that filtered with the same condition defined the baseline of the correlation. Significant correlations corresponded to cases where two consecutive binned data points of the CCG of one cluster pair exceed the means plus five times of the s.d. of the baseline. In such scenarios, the correlation strength of the cluster pair represents the peak amplitudes of CCG normalized by the baseline mean value, and the lag represents the bin location of the peak.
In vivo recording in mice
Mice were anesthetized using vaporized isoflurane (3% for induction and 1.5–2% for maintenance) and positioned in a stereotaxic apparatus designed for small animals (David Kopf Instruments). A craniotomy (~1.0 mm diameter) was made over the somatosensory cortex at anterior–posterior coordinates 0.0 to –1.5 mm and medial–lateral coordinates ±2 mm. For reference, a stainless-steel screw (18-8 button head Torx screw, #90910A600, McMaster-Carr) with a silver wire lead (0.375 mm diameter, #64-1320, Warner Instruments) was implanted in the contralateral occipital bone.
A 64-channel microelectrode array (4 shanks spaced 250 µm apart, 16 electrodes per shank at 50-µm vertical intervals; model A4x16_5mm_250-177, NeuroNexus Technologies) was then inserted through the craniotomy, targeting a depth of 750 µm for the deepest electrode on each shank. The array was interfaced with an analogue-to-digital amplifier (#C3325, Intan Technologies) mounted to the stereotaxic frame.
Pharmacological tests, optogenetic stimulations, neural circuit manipulations and disease phenotypes
Evaluations of pharmacological responses included exposure to 4-AP (Sigma-Aldrich, 275875-1G) at a concentration of 100 μM, which evoked immediate synchronous activity in spinal organoids. Lidocaine, introduced at a concentration of 2 mM (Sigma-Aldrich, L7757-25G) and then washed out after 20 min, inhibited calcium dynamics and disrupted connectivity. TTX at a concentration of 100 μM produced notable inhibitory effects, reducing overall activity and decreasing the number of active channels. Optogenetic transfection used Hues 3 ESC-derived spinal organoids at least 40 days old and approximately 1.5 mm diameter in size. Transfection was performed at a multiplicity of infection of 20, assuming 30,000 live cells, using a lentivirus from VectorBuilder (Vector ID: VB900122-3326aya) as the basis for studies of optogenetic activation. The EF1A promoter of the lentivirus delivered channelrhodopsin-2 to the cellular genome, resulting in EGFP expression in transfected cells. After 24 h of incubation, fully changing the cell culture media removed the lentivirus. Feeding of the virally transfected organoids occurred every Monday, Wednesday and Friday. Integration of transfected organoids with interfaces occurred 4 days after transfection, with optogenetic activity induced and recorded 10 days after transfection. For neural circuit manipulation, application of two clinical doses (2 U) of BTX (OnabotulinumtoxinA, AbbVie) diluted in 2 ml biomedia to neural organoids formed a concentration of 1 U ml−1. Washing removed BTX after incubation at 37 °C for 48 h. Recordings were made before and on each day after exposure to BTX. Disease modelling used glutamate (Thermofisher, J63424.14) at a concentration of 25 μM, followed by 2 h incubation and washing. Monitoring the neural activity captured the responses in all the tests above before the dosing, and at specific time after dosing (1 h for 4-AP, lidocaine and TTX, and every day for BTX and glutamate).
Statistical analysis
Statistical comparisons between experimental datasets were performed using a two-sided t-test. Statistical significance corresponded to P values of less than 0.05. Results shown in the figures use symbols to indicate statistical significance: NS (not significant), **P < 0.01 or *P < 0.001. These analyses used OriginLab.
Ethics statement
hiPS cell culture activities at the University of Illinois Chicago were conducted in accordance with Protocol 22-020, which was approved by the university’s Institutional Biosafety Committee. The WTC-11 line was derived from a donor who gave voluntary informed consent to a qualified professional. The cells used in this study were obtained from The Coriell Institute.
The human embryonic stem cell lines used in this study are National Institutes of Health-approved, and the human embryonic stem and hiPS cell research was conducted in compliance with the Shirley Ryan AbilityLab as well as Northwestern University’s Committee on Human Stem Cell Research registration requirements, which oversee the provenance, use and safekeeping of human pluripotent stem cells.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
First Appeared on
Source link






Leave feedback about this